This page provides help to the users of the TUPDB database on how to efficiently use this repository of target-unrelated peptides/motifs. For each module which is implemented in TUPDB, here we provide the figures and description on how to use that module. Three general modules are provided here.
Data browsing
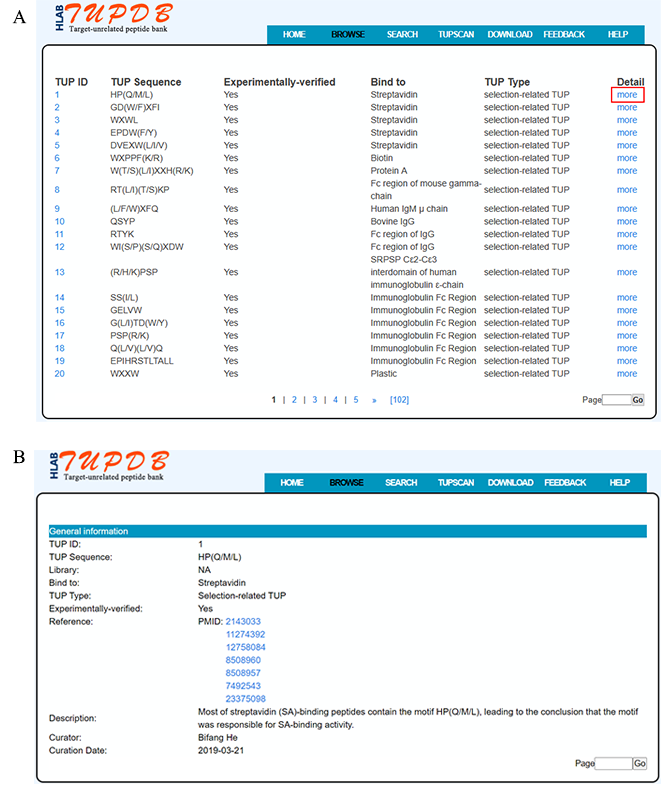
The TUPDB database can be conveniently browsed. By clicking 'BROWSE' at the top of TUPDB website, users firstly access a brief browsing table which displays fields including TUP ID, TUP Sequence, Library, Bind to, TUP Type and links guiding to the 'Detail' page. More detailed descriptions are provided in the 'Detail' page that can be reached by selecting 'more'.
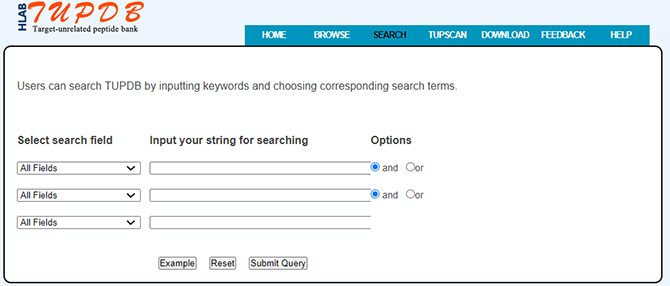
Data searching
TUPDB provides search results as a table in the search result page similar to browsing the database. Users can click 'SEARCH' to enter into the search interface and search TUPDB by inputting keywords and choosing corresponding search terms. Most fields can be used to search against the database.
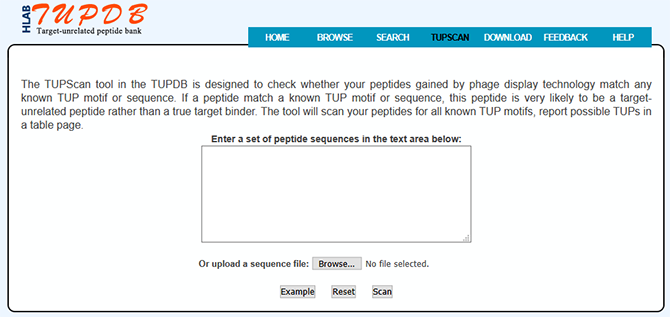
TUP matching
The TUPScan tool in the TUPDB is designed to check whether your peptides gained by biopanning match any known TUP motif or sequence. By clicking 'TUPScan' at the top of TUPDB website, users can enter into the interface of TUPScan.